[Ishikawa(Aogaku), Oshino, Yuzurihara]
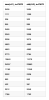
We tried using the latest Omicron (v4.1) installed by the IGWN conda and found that it's important to select the output format `hdf5` (not `root` format) to obtain almost the same triggers as the Omicron (v3.2). See the attached plots.
I updated the usage manual for Omicron (v4.1) to include an example and caution about the output format.
Detailed memo (for future workers)
-
For the previous Omicron (v3.2), we often used the output format of XML. But, the output format of XML was deprecated in the latest Omicron (v4.1). Omicron (v4.1) has two options for output format: root and hdf5. Initially we used `root` format and found that the number of triggers is significantly different between the `root` format in Omicron (v4.1) and `xml` format in Omicron (v3.2). This is because the clustering option will not be used in `root` format. (see the Omicron document) Due to this reason, I recommend using the `hdf5` output format instead of `root` in Omicron (v4.1).
-
We confirmed that the numbers of triggers are almost consistent between the `hdf5` format in Omicron (v4.1) and `hdf5` format in Omicron (v3.2). Actually, there is a slight difference. I guess the clustering method or something internal was updated over the version-up.
-
Currently, the Omicron with the old version (<<< v4.1) is running for the summary page. At some point in the future, we need to consider replacing the Omicron with a newer (or the latest) one.